Tackling Big Epidemics with Big Compute
Guest post by Kierste Miller, Data Scientist at Metabiota

At Metabiota, we are fascinated by infectious diseases and the way they spread. Epidemics pose an immense risk to the entire globe; however, they are notoriously challenging to forecast and monitor. Our team produces epidemic risk models for the insurance, commercial and government sectors to help address the challenge of quantifying this seemingly unquantifiable risk. Our end users are interested in knowing the probability of experiencing a certain level of human or financial loss due to infectious disease epidemics.
To assess the likelihood of loss, we produce in silico (i.e., performed via computer simulation) models that project plausible disease transmission events across the entire globe. For example, our simulators depict the potential spread of pandemic flu, as well as outbreaks akin to the 2003 SARS and 2014 West Africa Ebola events. We probabilistically model where disease emerges, how quickly it spreads, how many people it infects, and the resulting rates of healthcare utilization and mortality. Our clients are often interested in the costs associated with these events, so we couple disease spread models with financial models that quantify the economic impact and insurance claims related to outbreaks. Altogether, we create an extremely large set of simulated events that allows for the estimation of potential financial and human losses caused by disease epidemics.
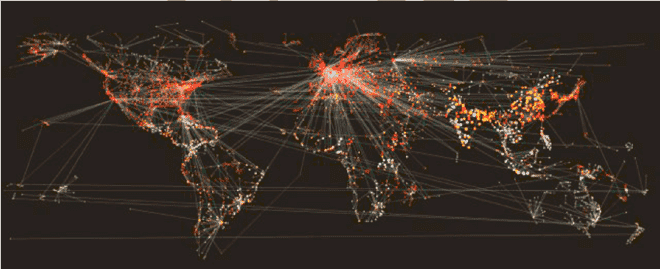
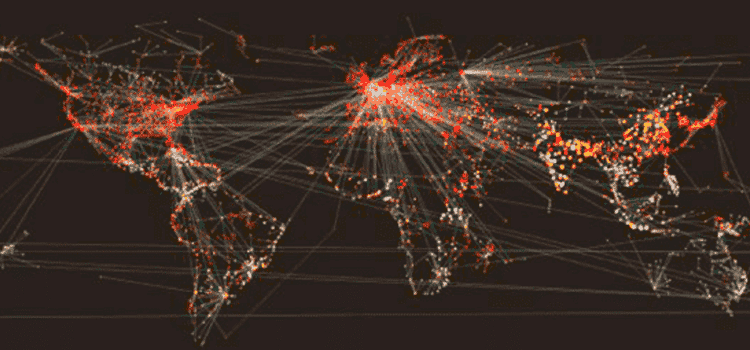
The map above illustrates how our models simulate the process of individuals traveling across the entire globe – from Paris, France to Paris, Texas for instance – on a daily time step. We model the probability of individuals moving across the network edges, as well as the probability of those individuals transmitting disease within each network node. There are millions of calculations occurring in a single epidemic simulation. Our models incorporate a huge amount of data and, naturally, we have significant big compute requirements.
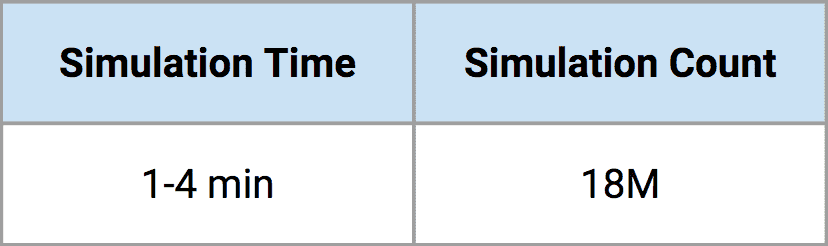
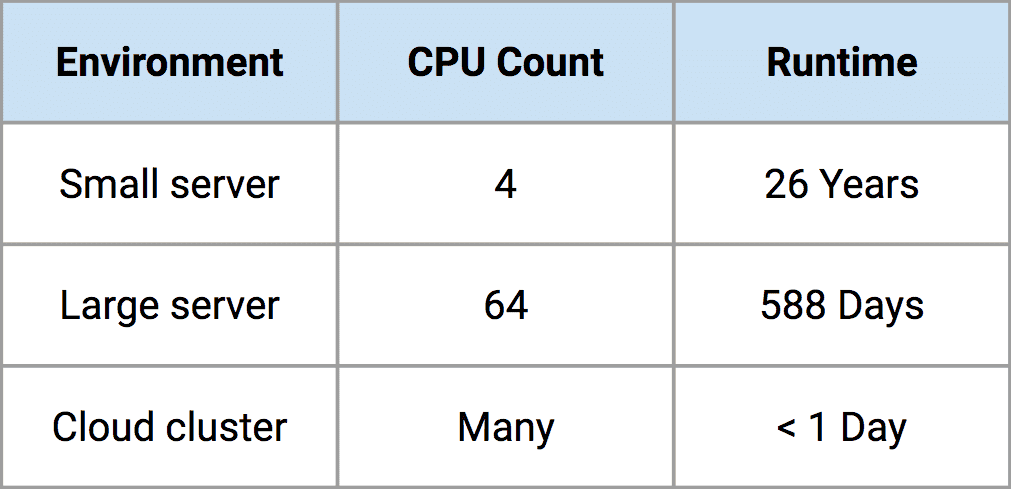
The table below summarizes our problem scope. Our clients are interested in the probability of rare events, so we actually simulate millions of infectious disease outbreaks to look at the entire range of plausible outcomes. Each simulation incorporates the transmission of disease within and between all nodes around the globe and takes approximately one to four minutes to complete. Given that we want to run millions of simulations to estimate the breadth of epidemic outcomes for a single pathogen, we are talking about a very large computing task. As you can see from the table, the set of simulations for one pathogen family would take about 26 years on my own laptop! That’s clearly not helpful, so instead, we use Rescale’s cloud computing platform to speed things up. On the Rescale platform, we are able to reduce 26 years of computing for a single pathogen down to approximately one day’s work. At the end of a single day, we have profiled a single pathogen’s risk using over 18 million simulations that capture a wide range of plausible scenarios.
We use cloud computing on Rescale because it enables us to do our computing faster, cheaper and more collaboratively. Rescale’s platform is scalable – we can run millions of global epidemic simulations across thousands of servers, and we are able to leverage big compute resources on a huge scale in a cost-efficient manner. We can collaborate securely on the platform with our partners around the world to build state-of-the-art infectious disease risk models and easily control access to the data. It’s a wonderful technological achievement to be able to cheaply and quickly simulate the complexities of infectious disease spread across the globe.
This is just one particular way in which the life sciences benefit from the big compute capabilities of today. There are billions of dollars and human lives at stake when infectious diseases spread – like the Ebola outbreak across West Africa in 2014 – and we are building tools to garner critical information to increase resilience to epidemics thanks to the simulation results that we obtain via Rescale.
About the Author
Kierste Miller is a data scientist at Metabiota focusing on modeling infectious diseases and specializes in computational biology, epidemiology and catastrophe modeling. She is a member of a team that builds global epidemic risks estimation products for insurance, commercial, and government sectors.