CONVERGE
CONVERGE should be run using the following command:
converge
This will run CONVERGE on all cores specified in the hardware settings for the job.
Note: If you are trying to use Converge super licenses, make sure your command includes this flag as follows:
For CONVERGE versions 2.x use:
converge -a super
For CONVERGE versions 3.x use:
converge -S
We recommend using the Nickel or Emerald core type when running CONVERGE on Rescale.
Each fully-utilized Nickel nodeIn traditional computing, a node is an object on a network. ... More provides 16 cores and 640 GB of SSD storage. We therefore recommend running CONVERGE on a multiple of 16 cores when running on Nickel.
Each fully-utilized Emerald node provides 18 cores and 640 GB of SSD storage. We therefore recommend running CONVERGE on a multiple of 18 cores when running on Emerald.
Convergent Science sometimes provides its customers with custom builds of CONVERGE which are not available to the public. The correct command depends on whether you are running CONVERGE 2 or CONVERGE 3.
Running a custom build of CONVERGE 2
You will need to upload the build executable file together with your input file. For custom builds to work, the executable has to be renamed to converge-2.X.X-<mpi-flavor>-CUSTOM (substitute the version number and mpi flavor). An example would be: converge-2.1.0-pmpi-CUSTOM. Please make sure that your custom build corresponds with the mpi flavor you have named, or your job will fail.
Once the renamed executable has been uploaded alongside with your input file, you need to run the job using the following command (on the Software Settings page):
converge -b CUSTOM
Your job will now run on your custom CONVERGE version.
Running a custom build of CONVERGE 3
Due to differences between the wrapper scripts for CONVERGE 2 and CONVERGE 3, custom builds of CONVERGE 3 require pointing directly to the executable. See an example of this below:
export LD_LIBRARY_PATH=${HOME}/work/shared:${LD_LIBRARY_PATH}
source
/program/converge-3.0.25/3.0.25/Convergent_Science/Environment/scripts/CONVERGE/CONVERGE-MPICH/3.0.25.sh
mpirun -np $RESCALE_CORES_PER_SLOT -machinefile $HOME/machinefile
converge-mpich-3.0.25_custom-cmv --super
Occasionally, you may need to restart a Converge job because it was stopped early or failed to complete. Rescale has built a feature that allows you to continue an incomplete job from the last written restart by following the procedure below:
- In the Results page of your job, click the Select all # files button as it appears on the top right corner. An example is shown below:
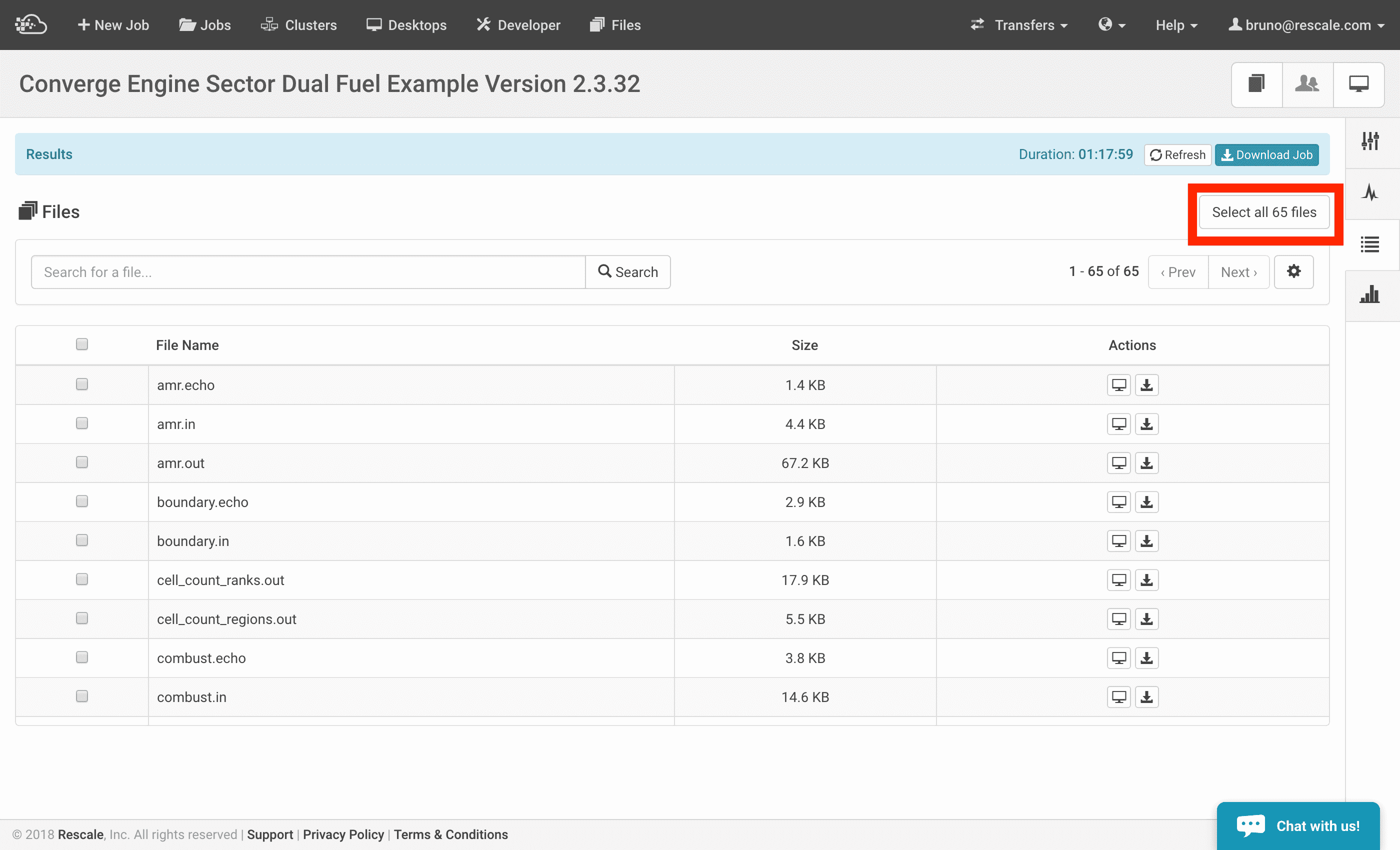
- Immediately, you will see options populated in a drop-down menu. Select Clone this job with selected files as input files.
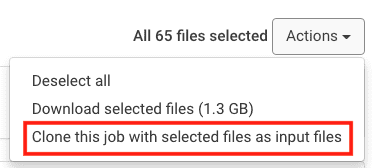
- The platform will perform such action, it will clone a new job and include all results files as input files. If you wish to maintain the same hardware settings (e.g. coretypePre-configured and optimized architectures for different HPC... More, and number of cores), submit your job. Otherwise, you may also wish to update the hardware settings and then submit the job.
Keep in mind that if there are more than 1 restart files as input files, Converge will choose the latest restart file and continue the analysis from that point
