Scaling Genetics Research with Cloud Computing
Advances in technology in the past decade have decreased the cost of sequencing the human genome significantly. With lower costs, researchers are able to perform population studies for various disorders and diseases. To perform these population studies, they will need to sequence thousands of patients and as a result, generate a significant amount of data with 70-80 GB files for each sample. After sequencing these patients, they will need to analyze the data with the goal of determining the cause of these genetic diseases and disorders.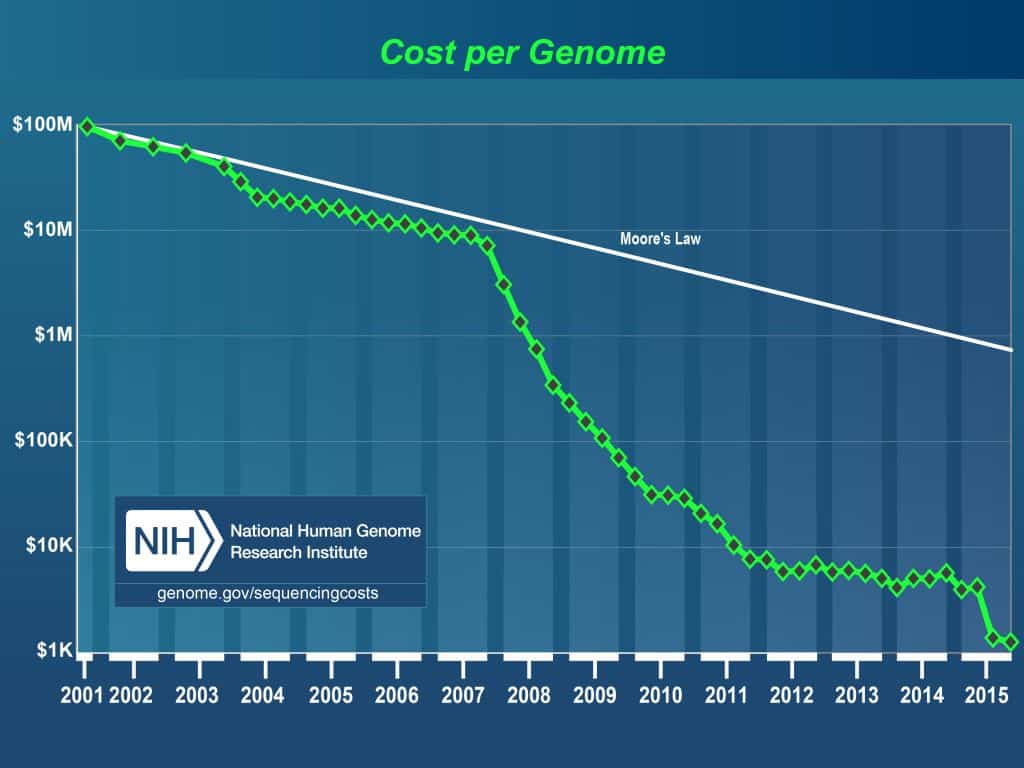
Using the generated data, end users of HPC systems run various analysis workflows and draw their conclusions. On-premise systems have many limitations that affect their workflow. These limitations include rapid growth of on-premise storage needs, the command line user interface, and full utilization of compute resources. Taking into consideration the logistics of expanding a storage server (purchase order, shipping, and implementation), end-users could be waiting over a month until they can start using their purchased storage. In academic institutions, graduate students (usually coming from a biology background) run analysis workflows on generated data. Most of the time, these students have never seen a command line prompt before. As a result, students must learn UNIX basics and HPC scheduler commands before they can even start running their analyses. Full resource utilization delays queued jobs from being scheduled onto compute resources. This limitation affects researchers greatly when research paper submission deadlines need to be met.
Managing an on-premise HPC system creates a high workload on an organization’s IT team. IT workers must constantly analyze their cluster usage to optimize the performance of the system. This optimization includes tuning their job scheduler to run as many jobs as possible and capacity planning for growing resources in their data center. Depending on the country, clinical data must be retained for a predetermined number of years. To address this constraint, IT workers must also implement and manage an archive and disaster recovery system for their on-premise storage in the event that researchers’ studies are audited.
These end-user limitations can be resolved and the workload on IT can be reduced through the use of cloud services like Rescale. By using Rescale storage, end-users pay for exactly what they use and are able to use their desired amount of storage instantly. Users are able to set policies using Rescale storage to automate archiving data. Through the use of our cloud storage solutions, data redundancy is as simple as clicking a checkbox. What’s more, researchers who adopt a cloud-native compute environment will be best-positioned to fully realize the benefits of the cloud by avoiding file transfer bottlenecks. Researchers should first move their data to the cloud, then incrementally push sequenced data to the cloud. The one-time cost of this transfer pays off in the long-run—the cloud offers researchers a highly flexible, scalable, long-term solution that puts unlimited cloud compute resources at researchers’ fingertips so they can always meet their deadlines.
Rescale’s cloud platform enables researchers to increase the speed and scale of their genetic analyses. As a result, they are able to obtain qualitative/quantitative data needed to publish their research papers. Discoveries made in these research papers will advance personalized medicine and eventually will be applied in a clinical setting with the goal of improving an individual’s health, quality of life, and creating a better world.
If you are interested in learning more about the Rescale platform or wish to start running your analysis in the cloud, create a free trial account at rescale.com.