LAMMPS Examples
LAMMPS, or Large-scale Atomic/Molecular Massively Parallel Simulator, is a molecular dynamics simulator. LAMMPS has potentials for solid-state materials (metals, semiconductors) and soft matter (biomolecules, polymers) and coarse-grained or mesoscopic systems. It can be used to modelA numerical, symbolic, or logical representation of a system... More atoms or, more generically, as a parallel particle simulator at the atomic, meso, or continuum scale. It computes using the solution using spatial-decomposition techniques and hence is able to parallelize well across multiple processors. On parallel computers, LAMMPS uses spatial-decomposition techniques to partition the simulationSimulation is experimentation, testing scenarios, and making... More domain into small 3d sub-domains, one of which is assigned to each processor.
On Rescale, LAMMPS simulations can be run easily across multiple cores and the results can be managed conveniently.
LAMMPS Basic Example
This example is a simulation of a rhodopsin protein in a solvated lipid bilayer.
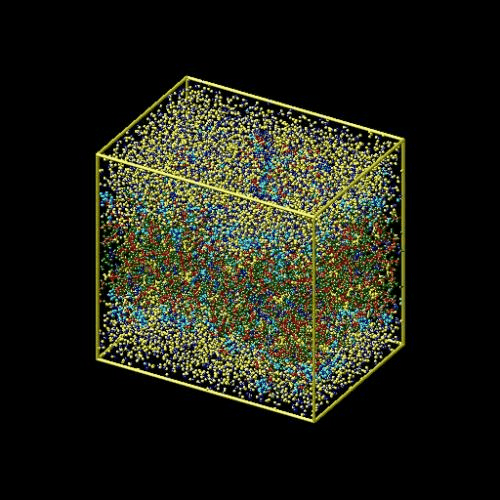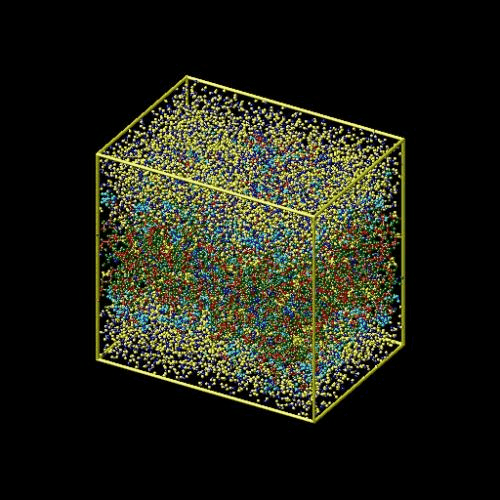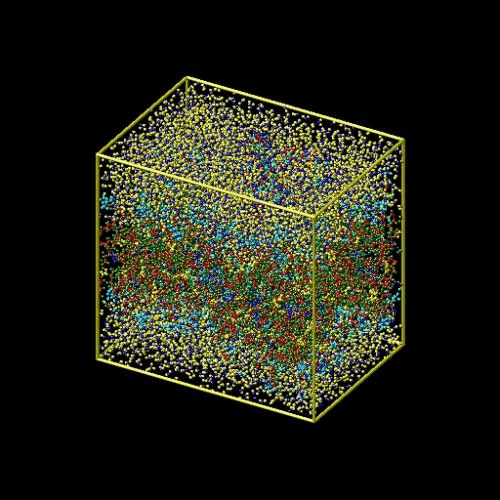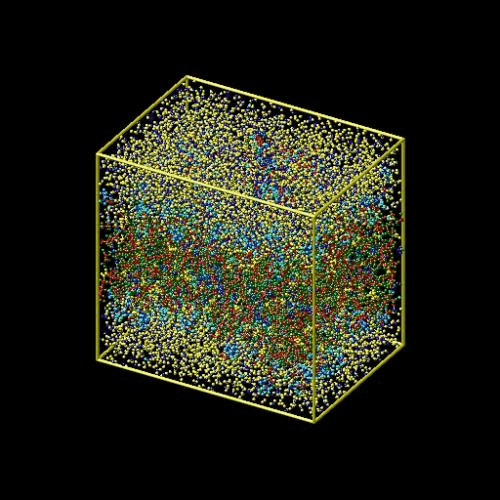
Post processing visualizationVisualization is the representation of complex scientific or... More of results
| Simulation Code | LAMMPS Mar 2019 |
| Analysis Type | Molecular dynamics |
| Description | All-atom rhodopsin protein in solvated lipid bilayer with CHARMM force field, long-range Coulombics via PPPM (particle-particle particle mesh), SHAKE constraints. |
| Suggested Hardware | Emerald / 2 cores |
| Command |
mpirun lmp_rescale -in in.rhodo |
| Estimated Run Time | 10 minutes |
| Previous versions | Import Job Setup Lammps Aug 2015 |
—
