Running NWChem on Rescale
Overview
Nwchem is an open source software developed in Pacific Northwest National Laboratory (PNNL). Originally designed for parallel architectures and scalability to 10,000’s of processors (part even to 100,000).
NWChem provides many methods for computing the properties of molecular and periodic systems using standard quantum mechanical descriptions of the electronic wavefunction or density. Its classical molecular dynamics capabilities provide for the simulationSimulation is experimentation, testing scenarios, and making... More of macromolecules and solutions, including the computation of free energies using a variety of force fields. Broad range of molecules, including biomolecules, nanoparticles and heavy elements Electronic structure of molecules (non-relativistic, relativistic, ECPs, first and second derivatives) can be modeled using NWChem.
Introductory Tutorial
In this example you will calculate the Electrostatic potential and the magnitude of the electric field for NaCl using NWChem.
This tutorial will introduce you to submitting NWChem jobs into the Rescale platform. An input file is already created on the local computer and we will show you how to start a Rescale job, submit, and transfer the results back to your computer.
Configuring Your Job
Create Job
To start up the Rescale platform, go to the platform.rescale.com and you can log in using your account information.
From the main screen of the platform, click on the +Create New Job button at the top left corner of your screen. Feel free to rename the job by clicking on the pencil icon next to the job name (“Untitled Job”) at the top of the page.
This is the first step of submitting your job.
Input File
The next step is to upload the input file.
You can either upload the input file from your local computer or if you already have it on the cloud, you can upload it using the cloud storagea simple and scalable way to store, access, and share data o... More.
For this tutorial you will want to upload the NaCl.nw file you downloaded at the start of the tutorial. On completion you will see your input file setup page look like that shown below:
Software Settings
The next step is to add the desired software, you can search for or directly select the tile for NWChem. For the purpose of this tutorial, NWchem 7.0.2 is used.
Under the Analysis options tab you are able to choose the preferred version and appropriate command line.
The analysis execution command is specific for each software package and each input file being used. For this input file you will need to add the following:
mpirun -np $RESCALE_CORES_PER_SLOT nwchem NaCl.nwNWChem is an open source software, so you don’t need to specify any type of license.The completed Software Settings should look like those in the image below:
Hardware Settings
Now we can select the type and number of cores to run on in the Hardware Settings page.
For a basic job, there are three hardware settings to edit: CoretypePre-configured and optimized architectures for different HPC... More, Number of Cores, and Walltime.
In this case, select 2 or more for the Number of Cores (1), 6 or more Hrs for the Walltime (2), and Emerald for the Corean individual processing unit within a multicore processor o... More Type. Rescale offers On-Demand Economy and On-Demand Priority core type options. You can find more information on these options here.
There is no need to specify any Post Processing options for this tutorial. To continue to Review, click Next from the Post Processing screen.
After reviewing your selections, your case should now be ready for batch submission at this point.
Review
The Review step shows you a summary of your job prior to submission. To submit the job click the blue Submit button in the top right corner on any of the job configuration pages or in the middle of the Review page.
Alternatively, instead of submitting the job, you can elect to Save the problem setup to be run at some later time.
Status
Now you can monitor the progress of your job from the Status tab. You can monitor the status of your job and get information about your clusterA computing cluster consists of a set of loosely or tightly ... More in real-time. And also you can see the output “process_out.log in live-tailing.
Because this analysis is entirely run in the cloud, feel free to close your browser window or shut down your computer. You can check on the progress at any time by logging into Rescale and clicking on the Jobs tab. You will receive an email notifying you when the job is completed. A guide on “Monitoring Status” on Rescale is found here.
Results
Once the job has completed its run, you can Download all of the output files from the Results page. You can also launch a Desktop and attach this job to perform post-processing.
A guide on Managing your Job Results on Rescale is found here.
Configuring Your Desktop
Once the job is completed, follow these steps to view results on a Rescale Desktop:
Set up a Desktop session:
- Under the Workstations tab, select the Classic Desktop.
- From the main screen of the platform, click on the +Create New Desktop button at the top left corner of your screen. Feel free to rename the desktop by clicking on the pencil icon next to the desktop name (“Untitled Desktop”) at the top of the page. Since Rescale’s platform saves all of your desktops , it is recommended that you name it something specific so that you will be able to find it again later.
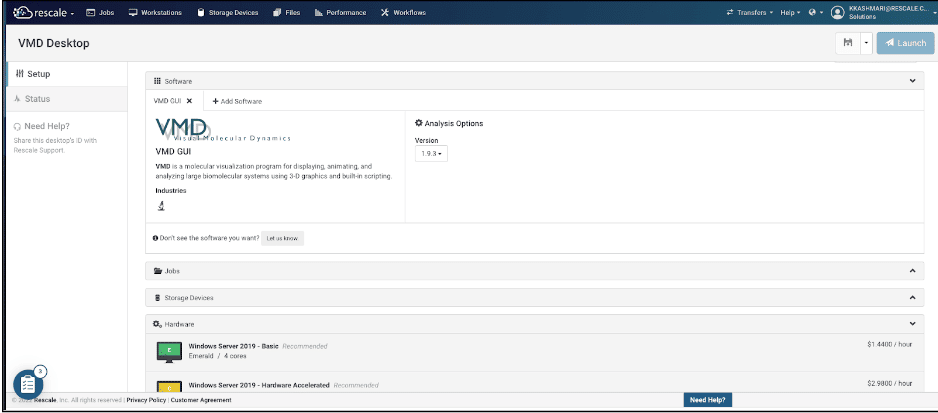
- In the next step, you can configure the software you need.
- For the purpose of this tutorial ,VMD GUI is selected. under the Analysis option tab you can choose your preferred version which is considered 1.9.3 for the purpose of this tutorial.
- Under the jobs section , you can attach the specific job that you want to use on the Rescale desktop.
- For the hardware it is recommended to use the “Windows Servera server is a computer program that provides services to oth... More 2019 – Hardware Accelerated”, which is well suited for the purpose of this tutorial.
- Now you can select launch on the top right corner of the platform, and wait a couple of minutes for the desktop to connect.
Once the desktop is created, select the Connect button on the top right corner of the platform:
Now that we are connected to the system, we can launch the software by double clicking it and load our results. Any job that is attached to the desktop is accessible through a folder on the desktop.
To visualize the Electrostatic potential (ESP) surface map:
- On the VMD top bar, select File and choose the New molecule.
- Find the esp-pad.cube file from the attached_job file
- In the next step go to the Graphics → Representations and change the Coloring Method to Volume and the Drawing method to Isosurface.
- Change the Isovalue to 0.1.
- Change the Draw to Solid Surface and the Show to Isosurface.
- The final setup page look like that shown below:
- In the next step, go to the Graphics → Colors , in the Color Scale tab change the color type to RGB:
- Now go back to the Graphical Representations → Trajectory and change the color scale data range from -0.00 to 0.10.
The ESP map of NaCl should approximately look like this:
